Summary
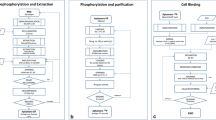
Prototype computer software for a Cell Culture Laboratory Management System (CCLMS) has been developed to relieve cell culture specialists of the burden of manual recordkeeping. Conventional data archives in cell culture laboratories are prone to error and expensive to maintain. The reliance upon cell culture to provide models for biochemical and molecular biological research serves to magnify errors at great expense. The CCLMS prototype encapsulates a modular software application that manages the many aspects of cell culture laboratory recordkeeping. A transaction-based database stores detailed information on subcultures, freezes and thaws, prints waterproof labels for culture vessels, and provides for immediate historical trace-back of any cultured cell line. Linked database files store information specific to an individual culture flask while removing redundancy between similar groups of flasks. A frozen cell log maintains locations of all vials within any type of cryogenic storage unit, locates spaces for newly frozen cell lines, and generates alphabetical or numerical reports. Finally, modules for maintaining cell counts, user records, and culture vessel specifications to support a comprehensive automation process are incorporated within this software. The developed CCLMS prototype has been demonstrated to be an adaptable, reliable tool for improving training, efficiency, and historical rigor for two independent cell culture facilities.
Similar content being viewed by others
References
Blessing, J. A.; Bundy, B. N.; Reese, P. A.; Priore, R. L. Experience with the use of generalized data base management systems in cooperative group clinical trials (a project of the Gynecologic Oncology Group). Control Clin. Trials 8: 60–66 1987.
Cataldo, R., Jr. Selecting database software: analyzing your needs. Top. Hosp. Pharm. Manage. 9: 9–16; 1989.
Dale, R. F.; Midwinter, M. J. Use of database management system by surgeons to produce operation notes. Ann R. Coll. Surg. Engl. 78: 272–275; 1996.
Doyle, A. The cell: selection and standardization. In: Dosle, A.; Griffiths, J. B., ed. Cell and tissue culture: laboratory procedures in biotechnology. 1st ed, London, UK: Wiley; 1998: 332.
Dreesen, R. G. A computerized database management system for the radiology research laboratory. Radiol. Technol. 58: 33–35; 1986.
Elmasri, R. The relational model and relational algebra. In: Navathe, S. B., ed. Fundamentals of database systems. 2nd ed. Vancouver, Canada: Benjamin/Cummings Publishing; 1994: 138–184.
Essin, D. J. Experimental identification of technical and database factors that can affect the success of clinical computer systems. Int. J. Clin. Monit. Comput. 5: 207–215; 1988.
Falconer-Smith, J. F. Assay Finder—an online database for clinical chemistry assays. Clin. Chim. Acta 278: 95–102: 1998.
FileMaker: File Maker Pro, version 5.0. Cupertino, CA: FileMaker; 2000.
Fischer, A. E.; Grodzinsky, F. S. Control structures. In: Zobrist, B.; Dworkin, A.; Herbst, N. ed. The anatomy of programming languages. New Haven, CT: Prentice-Hall: 1993: 248–285.
Fleege, J. C.; van Diest, P. J.; Baak, J. P. Quality control methods for data entry in pathology using a computerized data management system based on an extended data dictionary. Hum. Pathol. 23: 91–97; 1992.
Food and Drug Administration. Code of federal regulations Washington, D.C.: National Archives and Records Administration; 1999. Title 21, Chapter 1, Part 58.
Freshney, R. I. Culture of animal cells: a manual of basic technique. In: Freshney, R. I., ed. Chichester, NY: Wiley: 2000: 174, 191, 303–305
Hahn, P. F.; Lee, M. J.; Gazelle, G. S.; Forman, B. H.; Mueller, P. R.. A simplified HyperCard data base for patient management in an interventional practice: experience with more than 4000 cases. Am. J. Roentgenol. 162: 1443–1446; 1994.
Hasman, A.; Silkens, R. M.; Zinken, P.; Karim, A. B.; Westerman, R. F. ADAMO revisited: an interpretative review of a data management system. Int. J. Biomed. Comput. 23: 21–32; 1988.
Hoeck, W. G. InfoTrac TFD: a microcomputer implementation of the Transcription Factor Database TFD with a graphical user interface. Comput. Appl. Biosci. 10: 323–327; 1994.
Horrooks, I.. Constructing the user interface with statecharts. Reading, MA: Addison-Wesley; 1999: 253.
Lavappa, K. S.; Macy, M. L.; Shannon, J. E. Examination of ATCC stocks for HeLa marker chromosomes in human cell lines. Nature 259: 211–213; 1976.
Linzmayer, O. Dymo: Labelwriter. San Francisco, CA: Dymo-CoStar; 1999.
Long, W. J.; Shmihluk, K. E.; Gruver, P. E.; McGuire, W. R.; Palombo, A.; Emini, E. A. A voice-entry data management system: application in monoclonal antibody research. Comput. Methods Programs Biomed. 23: 211–216; 1986.
Lun, K. C. New user interfaces. Int. J. Biomed. Comput. 39: 147–150; 1995.
Mather, J. P.; Roberts, P. E.. Introduction to cell and tissue culture: theory and technique. In: Bunbar, B. S., ed. Introductory cell and molecular biology techniques. 1st ed. South San Francisco, CA: Plenum Press; 1998;1–87.
Meaders, R. A.; Sullivan, S. M. The development and use of a computerized database for the evaluation of facial fractures incorporating aspects of the AAOMS parameters of care. J Oral Maxillofac. Surg. 56:924–929; 1998.
Moritz, T. E.; Ellis, N. K.; VillaNueva, C. B., et al.. Development of an interactive data base management system for capturing large volumes of data. Med. Care 33:OS102-OS106; 1995.
Nakagawa, A. S. LIMS: implementation and management. In: McDonald, ed. London, UK: The Royal Society of Chemistry: 1994.
Nayler, O.; Stamm, S. Sciencel LabDatabase: a computer program to organize a molecular biology laboratory. Bio Techniques 26: 1186–1191; 1999.
Paul, J. Cell and tissue culture. Denver, CO: Livingstone, 1959.
Paul, J.. Cell and tissue culture. In: Paul, J., ed. Glasgow, UK: Longman: 1975.
Raskin, J. The humane interface: new directions for designing interactive systems. Reading, MA: Addison-Wesley; 2000: 233.
Russell, C. K.; Gregory, D. M. Issues for consideration when choosing a qualitative data management system. J. Adv. Nurs. 18: 1806–1816; 1993.
Seuchler, S. A.; Skolnick, M. H. HGDBMS: a human genetics database management system. Comput. Biomed. Res. 21: 478–487; 1988.
Simone, F. P.. Cell storage facilities and inventory. In: Doyle, A.; Griffiths, J. B.: Newell, D. C. ed. Cell & tissue culture: laboratory procedures. Chichester, NY: Wiley; 1998: 1–3.
Smith, J. M. CELLBASE: a database management system for the control of cell line storage. Comput. Appl. Biosci. 4: 408; 1988.
Timmers, T.; Pierik, E.; Steenbergen, M.; Stam, H.; van Ginneken, A. M.; van Mulligen, E. M.; Weber, R. F. ARIS: integrating multi-source data for research in andrology. Proc. Ann. Symp. Comput. Appl. Med. Care 1: 445–448; 1995.
Vierck, J. L.; Byrne, K.; Mir, P. S.; Dodson, M. V. Ten commandments for preventing contamination of primary cell cultures. Methods Cell Sci. 22: 33–41; 2000.
Vierck, J. L.; Dodson, M. V. Interprelation of cell culture phenomena. Methods Cell Sci. 22: 79–81; 2000.
Wear, L. L.; Pinkert, J. R. Reliable computer systems. J. AHIMA 64: 30–32, 34, 36; 1993.
Wu, C. L.; Wu, P. Shiau, A. L. A computerized management system for cell bank. Anal. Biochem. 275: 260–262; 1999.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Selznick, S.H., Thatcher, M.L., Brown, K.S. et al. Development and application of computer software for cell culture laboratory management. In Vitro Cell.Dev.Biol.-Animal 37, 55–61 (2001). https://doi.org/10.1290/1071-2690(2001)037<0055:DAAOCS>2.0.CO;2
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1290/1071-2690(2001)037<0055:DAAOCS>2.0.CO;2